
Protocol: CRISPR gene knockout protocol (Part 3): Single Cell Isolation and Positive Clones Validation
Among the CRISPR gene knockout protocols, the most commonly used and efficient method for CRISPR KO cell line generation is to use gRNA vectors to transfect into Cas9 stable cell lines, and obtain gene knockout cell clones through monoclonal isolation and screening. In this part, we will give a detail guidance of how to isolate a clone and validate it.
In case you haven’t read about our previous protocol, please use the following links. Vitro Biotech prepared a complete CRISPR KO protocol to guide you step by step to generate the KO cell line. This protocol includs:
CRISPR knockout cell line protocol (Part 1): gRNA Design and Vector Cloning
CRISPR-cas9 knockout protocol (Part 2): Cell Transfection
Part 3: Single Cell Isolation and Positive Clones Validation
Here is the part 3 of the CRISPR cas9 gene knockout protocol--Single Cell Isolation and Positive Clones Validation.

According to the preliminary test result of Part 2, if the cell line cannot form single-cell clones, it is recommended to continue the study with the CRISPR gene knockout cell pool. An efficient transfection plus a high quality cas9 expressing cell line can highly improve the KO effect.
Among the cas9 stable cell lines Vitro Biotech offers, these cell lines are tested unable to form clones. So right after cell transfection, you could perform PCR and sequencing to test the KO effect of the CRISPR cas9 gene knockout cell pool. If you have trouble interpreting the sequencing data, you could use ICE to analyze.
- SK-BR-3-CAS9
- Hepa 1-6-CAS9
- LLC-CAS9
- LLC-PK1-CAS9
- AR42J-CAS9
▍ Method 1: Flow cytometer isolation
1) Digest the gRNA transfected and screened cas9 expressing stable cells, adjust the cell concentration to 10^6/ml. Screened by flow cytometer, and then seed the cells into 96 well plates. The parameter is set to 1 cell/well. Single living cells are screened and seeded.
2) Continue to culture for about two weeks, during culturing, replenish or change the media in a timely manner. When the cell in the well grow to a cell clone, select the positive clones and expand them into each well of 24-well plate;
3) When the cells on the 24-well plate grow at 90% cell density, subculture each clone into the each well of 6-well plate according to the cell passage method, and a small amount of cells are collected for KO validation.
▍ Method 2: Limited dilution
1)Prepare a cell suspension, count the cells and perform series dilution until there are 10 cells per milliliter;
2)In 96 well plate, seed 0.1ml cell suspension per well, that is, each well contains 1 cell;
3)Continue to culture for about two weeks, during culturing, replenish or change the media in a timely manner.
Cas9 stable cell lines that can form single cell clones:
▍ CRISPR KO cell line validation
Sanger sequencing and WB are the most used methods for validating CRISPR cas9 gene knockout cell lines. Sanger sequencing is relatively simple, please refer to the “Preliminary experiment” of Part 2 for details; WB method is relatively intuitive, but requires reliable antibodies for this gene.
Here is the procedures for PCR and sequencing validation.
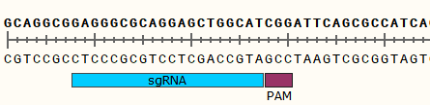
1) Extract gene DNA, amplify the genomic sequence near the knockout target by PCR, and identify positive knockout clones through sequencing.
2) Sequencing results show that a deletion of a fragment sequence near the target site (the number of the deleted base is not 3n) will result in a frameshift mutation in the gene as a positive clone.

3) The positive clones were further expanded in culture until the required amount of cells was obtained.

